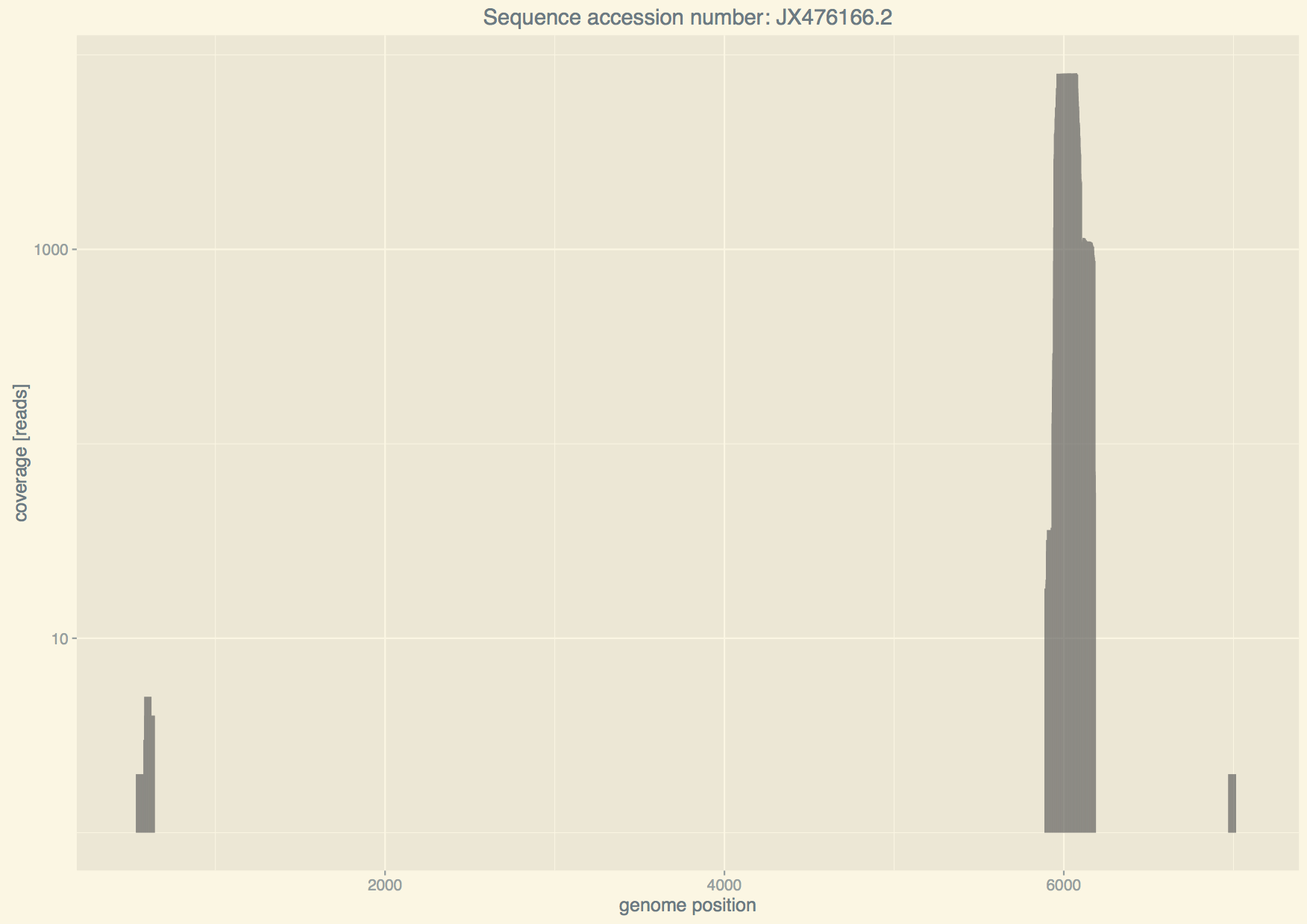
covplot: specify the genome to plot coverage
After a run of wolfpack we have a count of how many reads
in a sample are assigned to a given organism, but we might be interested to
know what fraction of the genome is covered by our reads because this would
give further evidence to the presence of the organism in our sample. In other
words, thirty reads from three different regions of the genome provide a stronger
evidence than thirty reads all from the same region. With covplot we can
easily create a plot of the coverage for an organism of interest.
Let's suppose we want to investigate sample AR-1_S1 in the directory
virmet_output_exp_01; we first look at the file listing organisms and reads
count
[user@host test_virmet]$ cat virmet_output_exp_01/AR-1_S1/orgs_list.tsv
organism reads
Human adenovirus 7 126
Human poliovirus 1 strain Sabin 45
Human poliovirus 1 Mahoney 29
Human adenovirus 3+11p 19
Human adenovirus 16 1
This seems to be populated by two viruses, some adenovirus and some polivirus.
Let's run covplot with --help to list the available options
[user@host test_virmet]$ virmet covplot --help
usage: virmet <command> [options] covplot [-h] [--outdir OUTDIR]
[--organism ORGANISM]
optional arguments:
-h, --help show this help message and exit
--outdir OUTDIR path to sample results directory
--organism ORGANISM name of the organism as reported in orgs_list.tsv file
If we want the coverage of reads on the genome of adenovirus we can run
[user@host test_virmet]$ virmet covplot --outdir virmet_output_exp_01/AR-1_S1 \
--organism "Human adenovirus"
covplot will perform the following steps:
- identify all mappings read-organism where the organism name starts with "Human adenovirus",
- identify the organism with the highest number of reads mapped,
- download the genome from Genbank and align all viral reads to it,
- compute the coverage, write it to
depth.txtand plot it.
The final result is a pdf file Human_adenovirus_coverage.pdf. Regarding point
1, it is important to note that giving "Human adenovirus" will chose the
genome with most hits from adenovirus 7 (top in the list). If one wants to see
how well viral reads would cover another adenovirus then one needs to give, e.g.,
--organism "Human adenovirus 3".